Spectroscopy
Structure Determination Case Study: Deer Tarsal Gland Pheromone
Last updated: October 31st, 2022 |
A Roadmap For Structure Determination of Organic Compounds
Male black-tailed deer emit a pheromone from their tarsal gland that attracts female deer.
Say you had a bunch of deer tarsal glands available and wanted to try to identify the active molecule from that gland.
How would you go about identifying the structure of the pheromone? What techniques would you use? How do you go from an “unknown” mixture to a “known” molecule? That’s what this post is about.
Table of Contents:
- The Four Stages of Natural Product Isolation
- Natural Product Isolation Case Study: Deer Tarsal Gland Pheromone
- A Roadmap for Structure Determination
- Analysis of an Unknown Compound
- Proposing a Structure Based On The Evidence
- Conclusion: Structure Determination
- Notes
1. The 4 Stages Of Natural Product Isolation
In January 1969, the Beatles released their “Yellow Submarine” single and performed their last live gig, Richard Nixon was inaugurated as president, and Brownlee et. al. published their seminal paper, “Isolation, Identification, and Function of the Chief Component of the Male Tarsal Scent in Black-tailed Deer” in the scientific journal Nature.
It’s that last part that we’re going to focus on today.

*statement may not be completely accurate
The Beatles Rooftop Concert 1969 London (HD)
Doesn’t Ringo look miserable?
Now on to something even more exciting: natural product isolation, which we’ve been going through over the past few posts.
You can think of the isolation of natural products (molecules) as having four main stages.
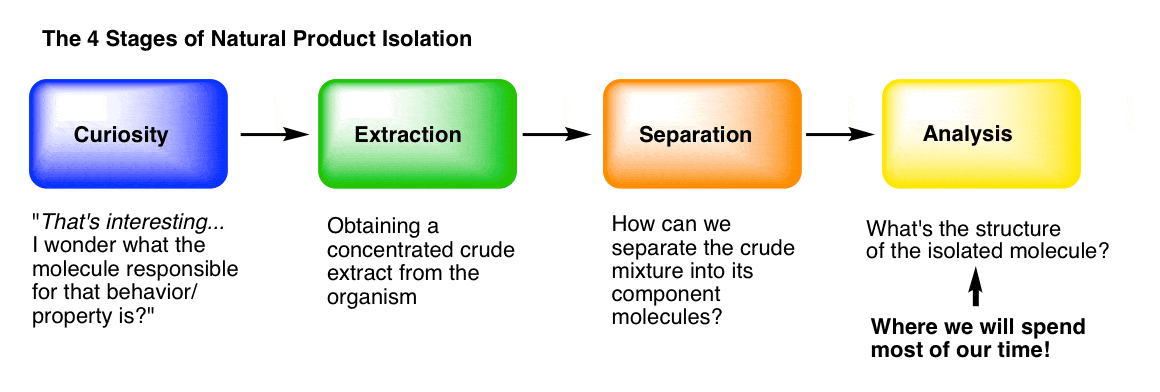
Stage 1: Curiosity. What makes “hot peppers” hot? What’s responsible for the painkilling properties of opium? How do insects find each other to mate, even from miles away? What gives vanilla that lovely aroma? [We went through some interesting molecules from nature in this post.]
Stage 2: Extraction. Following our curiosity, we need to find a way to get the molecules of interest out of the organism and into a flask. “Extraction” of the organism with an appropriate solvent is usually how we do it, which we covered here.
Stage 3: Purification. How can we separate a crude mixture into its components, so that we can determine which molecular species are present? We covered that in the last post, here.
Stage 4: Analysis. How do we determine the structure of a pure, unknown compound? This is what we’ll talk about today, and for a good long while in the future, because it’s not a small topic.
2.A Natural Product Isolation Case Study: Isolation and Analysis of Deer Tarsal Gland Pheromone
Since we’ll be covering analysis for a long time, why not start with something fun? In this post we’ll present a case study on how Brownlee et. al. began with an observation and a question (“Black-tailed deer smell each other’s tarsal areas a lot. What molecules are in the tarsal gland?”) and followed it through to determine the structure of the key pheromone below, which is an important signalling molecule for male black-tailed deer.
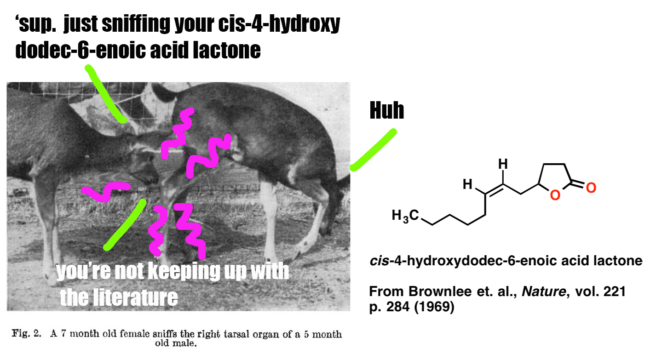
This study nicely illustrates the analysis of an unknown natural product. It’s also fairly simple, and you don’t need any knowledge of NMR [which we’ll cover in more detail later] to follow the logic of how the structure was obtained.
1. ‘Curiosity’ . Male deer of the genus Odocoileus contain a tarsal organ on their ankle which consists of scent glands and is covered with long tufts of bright, stiff hairs. Members of an established herd check each other’s tarsal tuft occasionally (once or twice an hour for each individual). When a strange female or strange male approaches the herd, the frequency of smelling tarsal turfs increases up to 6-16 times an hour (for a strange male). Fawns recognize their mother by sniffing her tarsal organ. When threatening each other, bucks often urinate on the tarsal tufts and rub them together simultaneously.
Clearly there’s a key molecule (or molecules) that provides this chemical signal. What’s its structure?

2. Extraction. Tarsal organs from male black-tailed deer killed in the same area during the same week were excised. The hair tuft was rinsed with petroleum ether (a mix of short-chain alkanes) and the collected extracts, after removal of petroleum ether, were distilled at 80-100 °C at reduced pressure to give a crude oil.
3. Separation. The extract was analyzed with gas chromatography (GC) which showed that the tarsal gland has many chemical components. The numbers at the bottom are in minutes: the major component (labelled 66) from the male tarsal gland at 16 minutes was chosen was chosen to be investigated. (Interestingly, the fraction labelled 26 at about 6 minutes was found to be the major component of female tarsal glands. )
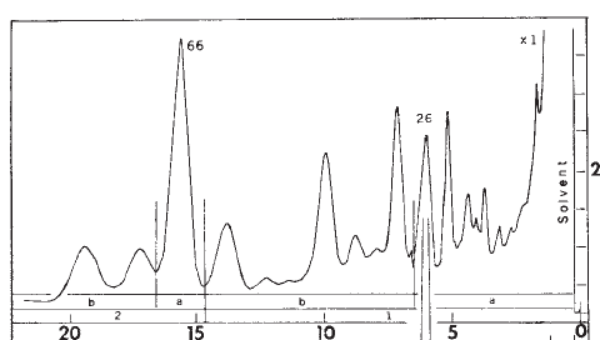
Isolation and analysis of this component was performed using “preparative” gas chromatography. Just for fun, I’m including full conditions in Note 1: Some of the jargon has been “decoded” to make it more accessible.
4. Analysis. Here’s where we really begin. The authors mention that a single buck organ yielded 10-80 micrograms (μg). 30 micrograms were used for analysis. That’s 1/30 of a milligram, folks: extremely impressive even by today’s standards.
Before diving in, let’s take a moment to survey the landscape.
Let’s say you have a pure sample of an unknown compound. Where do you start? What are the most important questions to ask?
3.A Roadmap For Structure Determination
I might humbly suggest the following, with the most important questions listed first.
- What’s the molecular mass? If you have the molecular mass (available from mass spectrometry), you can start to narrow down possibilities for its molecular formula.
- What’s the molecular formula? As we’ll see, in the absence of high-resolution mass spectrometry (not available in 1969) obtaining the molecular formula from the molecular mass still required some intuition and detective work.
- Once the molecular formula is known: how many degrees of unsaturation are present?
- What functional groups are present? Which functional groups can we rule out?
- What is the structure of the carbon skeleton? Is it linear? Branched? Does it contain rings? Multiple bonds? What about the placement of “heteroatoms” (O, N, S, etc. ) .
- Is there any stereochemistry to worry about? E/Z orientation of alkenes? Chiral centers?
These questions, although not comprehensive, are a good “road map” for the order of operations in determining an unknown structure. Notice how they start off with the big picture (molecular formula) and end up tying down small details (like the configuration of a stereocenter).
We’ll see that the structure determinination done by Greenlee et. al. roughly followed this general plan as well.
4.Analysis of An Unknown Compound
1. First, determine the molecular mass with mass spectrometry.
A small amount of the pure, unknown sample was injected into a mass spectrometer, which showed a molecular ion peak at 196. This means that the sum of the atomic masses* [Note 2] of the elements in the molecule add up to 196.
Is that enough to give us our molecular formula? Not quite. For example, using this website and restricting the search to molecules containing only C, H, and O (a reasonable restriction for hydrocarbons) shows at least six reasonable results for a mass of 196.
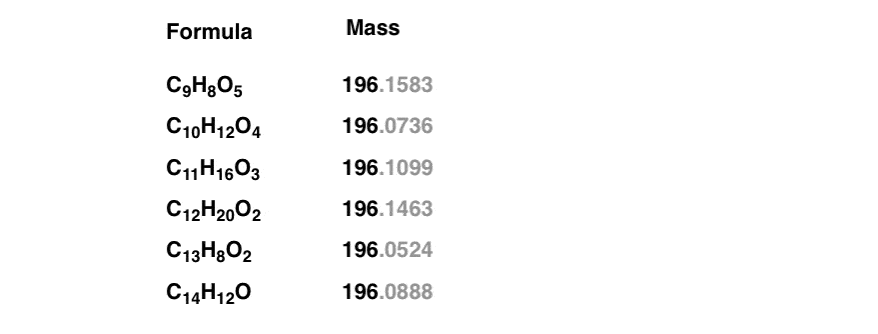
It’s a bit like a list of suspects in a police lineup. You know one of them is correct – but which one? How can we narrow this down further?
Note those extra decimal places in grayscale. In these more modern times, we commonly employ the technique of high resolution mass spectrometry, accurate to (at least) four decimal places. This can discern the slight differences in the mass of nuclei and help us immediately determine the molecular formula: a huge time-saver.
Since this technique wasn’t available to Brownlee et. al., some cleverness and intuition was required.
First, mass spectrometry showed that the base peak – i.e. the largest peak in the spectrum, corresponding to the most favoured fragmentation of the molecule – was at 85 , meaning that the parent molecule split into two fragments of masses 85 and 111. This information will be useful in a second – hold on.
2. Identifying key functional groups: Infrared (IR) spectroscopy
As we’ll see, infrared (IR) spectroscopy is an excellent technique for identifying certain functional groups. In particular, C=O and O-H groups vibrate at characteristic frequencies.
The IR spectrum of the unknown showed two key peaks:
- A strong peak at 1775 cm-1 , characteristic of a C=O stretch
- A strong peak at 1170 cm-1 , corresponding to a C-O stretch.
Taken together, the authors noted that these two peaks were strongly suggestive of a 5-membered lactone ring. They didn’t deduce this from first principles: this was inferred from comparison of their data with those from compounds isolated previously, much as police detectives might match fingerprints from a database. [Note 3]
[Like the “dog that didn’t bark“, almost as important as what peaks were found is what peaks weren’t found: no O-H, no N-H, no aromatic peaks, no triple bonds… ruling things out can be as clarifying as counting things “in”. ]
A “lactone” is a cyclic ester. So a “five membered lactone ring” looks like this.

Note the molecular weight of this fragment: 86. We noted earlier that mass spectroscopy showed that the molecule broke into two fragments, one of mass 85 and the other of mass 111.
We get 85 by taking that 5-membered lactone and removing a hydrogen. So that fragment of 111 is attached to the lactone somehow. But where?
When we cover mass spectrometry in future posts, we can devote more time to understanding fragmentation patterns. For now, a good rule of thumb is that molecules tend to break apart at sites which would form stable carbocations or radicals.
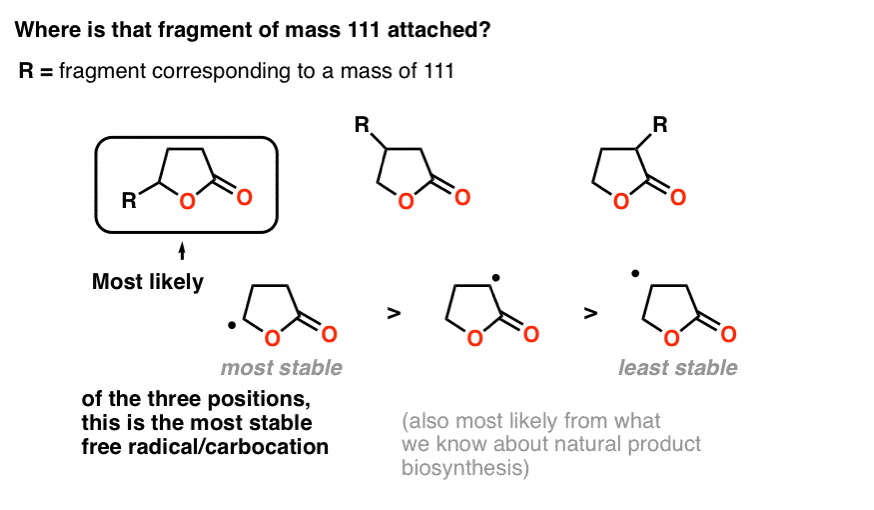
Attachment of the fragment to the carbon attached to the oxygen makes the most sense for two reasons.
First, fragmentation at that point would make for a relatively stable carbocation (or radical, if you prefer), which is a good way to think about likely points of bond cleavage in mass spectrometry.
Secondly – and we won’t go deeply into this – it is also simpler from a biosynthetic perspective, as it would give rise to a linear fatty acid instead of a branched one, and linear fatty acids are extremely common natural products.
3. Intuition provides a good guess for the molecular formula.
With the lactone identified, the next question becomes identifying that sidechain with molecular weight of 111. Now comes an intuitive leap: the authors banked on simplicity and made the assumption that it was a hydrocarbon of some kind, without oxygen.
A hydrocarbon with mass of 111 would correspond to C8H15.
Putting it together with the lactone, this gives us a molecular formula of C12H20O2 for the unknown compound.
4. Calculating the Degree of Unsaturation from the Molecular Formula
Now: once we have the molecular formula, we can use a nifty trick called the “degree of unsaturation” to determine the number of (double bonds + rings) in a molecule .
The formula for an alkane without double bonds or rings is CnHn+2 . Try it with methane (CH4) or ethane (C2H6) : the number of hydrogens is equal to twice the number of carbon atoms, plus two.
Every double bond or ring reduces the number of hydrogens in the molecule by two. So for C12 , we would expect [(2 x 12) + 2] = 26 hydrogens if there were no double bonds or rings (oxygen does not affect the number of hydrogens in any way). The actual formula is C12H20O2 , which gives us 6 fewer hydrogens. Since each double bond or ring removes two hydrogens, that means we have 6/2 = 3 double bonds/rings on the molecule, or 3 “degrees of unsaturation” .
Two of these three degrees of unsaturation are found in the lactone ring: the C=O bond (a double bond) and the ring itself. That leaves one degree of unsaturation for the side chain, C8H15.
Therefore, the side chain must contain either a double bond or a ring.
5. Determining the structure of the C8H15 sidechain : “Degradation” With Hydrogenolysis and Ozonolysis
The next question to answer was the structure of the C8H15 side chain.
Modern techniques such as NMR spectroscopy have greatly facilitated the process of determining the structure of a molecule. However, Brownlee et. al. didn’t have access to our sophisticated techniques. Instead, they used a relatively low-tech method to determine the structure of the side chain that has been used since time immemorial: a technique called degradation. Thankfully it’s a lot easier to explain for beginners than NMR.
Basically, “degradation” involves running chemical reactions on an unknown molecule to gain clues about its structure.
For instance if you have a molecule that you suspect has one or more double bonds, you can treat it with hydrogen and Pd/C (hydrogenation) and then examine the product to determine how many equivalents of hydrogen were added. For instance, if we took our unknown with a molecular weight of 196 and treated it with Pd/C and H2 to give a new product with molecular weight of 198 (observed by mass spectrometry) that’s a pretty good sign there’s a double bond in the molecule.
A related reaction to hydrogenation is “hydrogenolysis“, which we don’t really cover in introductory organic, but it involves exhaustive reduction of every functional group present to give an alkane, usually at high temperatures.
Brownlee et. al. wanted to determine if the side chain was linear or branched. So they subjected the unknown lactone to hydrogenolysis, and determined (by GC) that the main product was n-dodecane (C12H26), a linear alkane. Therefore, the side chain must be linear and contains a double bond (since it has one degree of unsaturation).
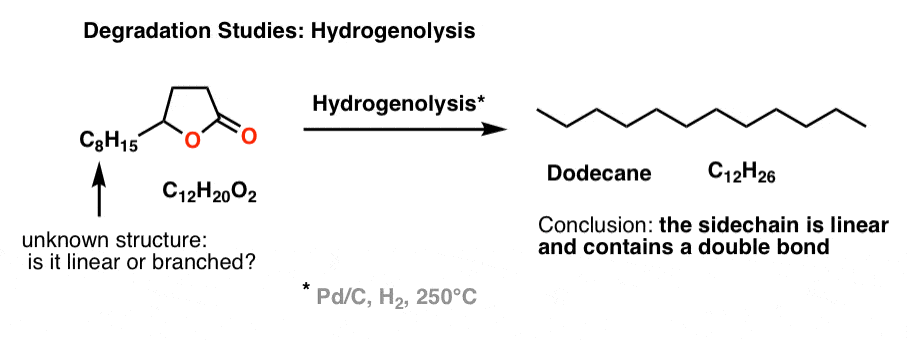
That led naturally to the next question: where is the double bond on the side chain?
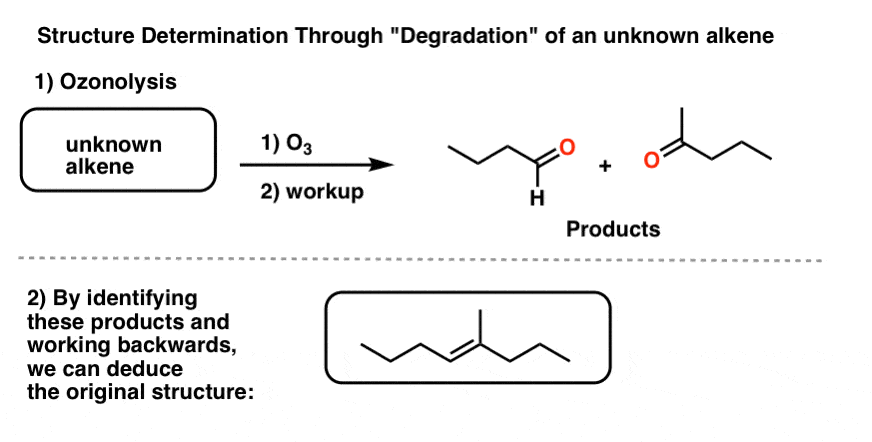
To address this question, another common reaction for degradation was rolled out: ozonolysis. Since ozone (O3) cleaves double bonds, we can take an unknown alkene, treat it with O3, and then analyze the products. Since we know how the reaction works in the forward direction, we can deduce the structure of the (unknown) starting material from the products.
Brownlee et. al. treated the unknown lactone (3 μg !!!) with O3 and then a reductive workup. They identified one of the products as hexanal by gas chromatography. Working backwards allowed them to determine the structure of the side-chain as shown.
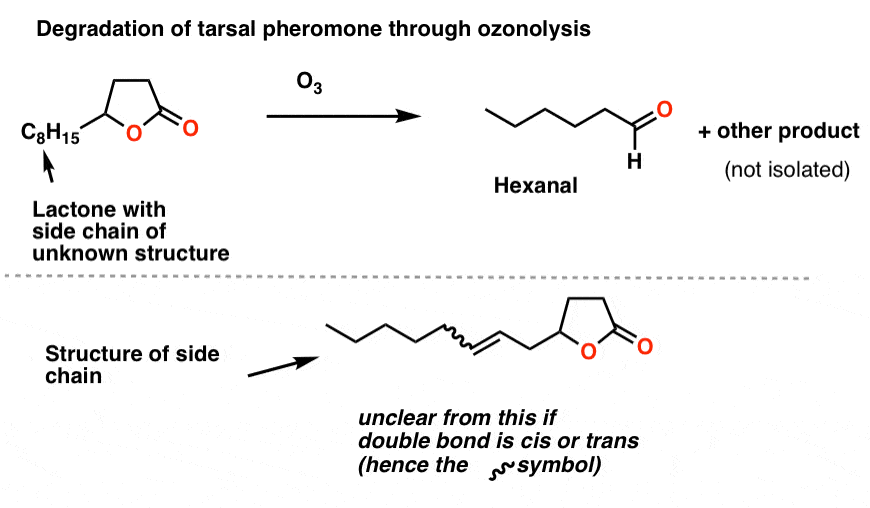
The last piece of the puzzle was the orientation of the double bond: cis or trans? Again, nowadays we’d use NMR for that, but they were able to deduce the orientation as cis from the absence of a band at 960 cm-1 [where trans alkenes typically have a peak]. [This is a subtle detail we don’t generally go into when we cover IR].
5.Proposing A Structure For The Pheromone Based On The Evidence
This gives the final structure of the pheromone:
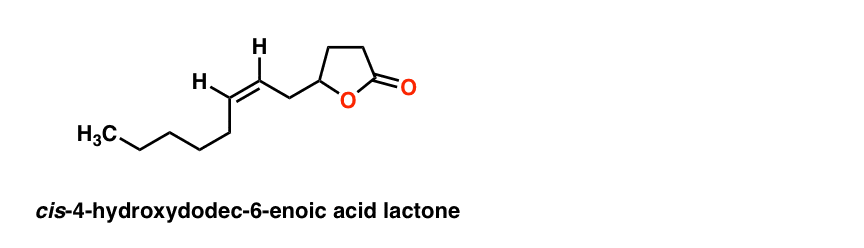
The final proof was obtained by comparison of the compound with an authentic sample provided by Unilever, which had identical characteristics in IR, MS, and GC spectra. Structure solved!
6. Conclusion
Hopefully this post has shown that determining the structure of an unknown compound really is like detective work.
It requires intuition, logical thinking, and attention to small details. That’s what’s so fun about it, IMO.
What’s even more rewarding in this case is that determining the structure of this molecule helped to peel back a bit of the mystery of how deer communicate with each other. It also leads to a host of other interesting questions: how is it made in nature? How did this process evolve? What other species communicate in this way? that we won’t have time to get into.
This case study has also introduced us to some of the key tools that chemists use to determine the structure of unknown compounds: IR spectroscopy, mass spectrometry, and (although only mentioned tangentially here) NMR.
It also introduced us to a framework for determining the structure of an unknown compound.
In the following posts, we’re going to move away from this side adventure on natural product isolation and start to discuss the individual components of structure determination.
We’ll start with this concept of the “degrees of unsaturation” . Next post!
Notes
Note 1. GC conditions, decoded.

- “Carbowax 20 M” refers to the stationary phase, a polymer of ethylene glycol (PEG) with average molecular weight 20,000 g/mol (hence the 20M).
- ‘Chromosorb W 60/80’ refers to the solid support that the Carbowax 20 M is loaded onto (i.e. chemically bonded with): diatomaceous earth, and the 60/80 refers to the “mesh size” (0.18 – 0.21 mm) which determines the surface area. Obviously a high surface area will improve separation, at the cost of requiring higher pressure to push solvent through.
- 1.5 m x 2.4 mm I.D. (inside diameter) refers to the size of the column
- 200°C is the temperature the column was heated to. Higher temperatures increase volatility of the gas and cause compounds to come out faster.
- 35 cm3 N2/min refers to the flow rate of nitrogen gas (the mobile phase) through the column.
- The “retention time” of 11.2 minutes means that it took 11.2 minutes for the desired compound to make it through the column under these conditions.
- “Fraction recut” means that the sample was somewhat impure, and they chose to send it through the GC again under slightly different conditions (using a different column, QF-1 instead of Carbowax – so far as I can tell from quick Googling, QF-1 is a fluorosilicone).
Note 2 * a quick digression on “mass”. Which “mass” should we use for each atom? We’re accustomed to using “molar mass” for most calculations, which is the mass of an atom averaged over an entire mole (6.02 x 1023 molecules). For instance the molar mass of carbon is 12.011 due to the presence of small amounts of 13C (1.1%). However, when an atom of carbon is measured by a mass spectrometer, it’s nonsensical to assign it a mass of 12.011 – it’s either 12 or 13! Hence, the molecular ion for a molecule represents the “mono isotope” form – that is, the mass of the molecule corresponding to each atom’s most numerous stable isotope. This is referred to as the “nominal mass” .
Note 3. Chemists of a certain age will remember these beasts from the reference section of the chemistry library:

Just WOW. You create the best site to learn and understand organic chemistry. As always, i am waiting on next posts.
Thanks Milosz – glad you enjoyed!